SE(3)-Transformer for molecular property prediction#
Authored by: Anuya Welling, James E. T. Smith, Diptorup Deb, Mukhil Azhagan Mallaiyan Sathiaseelan, Yao Liu, Phani Vaddadi, and Vish Vadlamani
The SE(3)-Transformer is a graph neural network that uses a variant of self-attention for 3D points and graphs processing. This model is equivariant under continuous 3D roto-translations, which means that when the inputs (graphs or sets of points) rotate in 3D space (or more generally experience a proper rigid transformation), the model outputs either stay invariant or transform with the input.
In the SE(3)-Transformer model, both training and inference are framed as regression tasks over a set of molecular properties. These include parameters such as μ, α, HOMO, LUMO, gap, R², ZPVE, U₀, U, H, G, and Cv, along with their atom-wise counterparts and rotational constants (A, B, C).
While the full physical interpretation of many of these quantities is best left to subject matter experts, this tutorial focuses on a few key electronic properties that are commonly used in molecular modeling. HOMO (Highest Occupied Molecular Orbital) refers to the highest energy level that is still occupied by electrons. LUMO (Lowest Unoccupied Molecular Orbital) is the lowest energy level that does not contain electrons and is available for occupation. The HOMO–LUMO gap, often simply called “the gap”, is the energy difference between these two orbitals and is an important indicator of a molecule’s electronic and chemical behavior.
In drug discovery, SE(3)-Transformer models predict molecular properties that matter for real biological behavior. A good drug must bind strongly to its target, avoid off-target reactions, and remain stable in the body while being reactive at the right site.
HOMO and LUMO capture this balance. They indicate a molecule’s tendency to donate or accept electrons, which influences how it reacts with protein amino acids and whether it could be toxic. The HOMO–LUMO gap acts as a proxy for reactivity – too small can mean instability and side effects, while too large can mean inactivity. Poor reactivity often leads to drug failure, even when binding looks promising.
For more information#
To find out more about DGL, SE(3)-Transformers, and AMD ROCm performance benchmarks, see the following blogs:
Graph Neural Networks at Scale: DGL with ROCm on AMD Hardware
Learn about DGL (Deep Graph Library), its design principles, and how it enables scalable graph neural networks on AMD hardware.DGL in the Real World: Running GNNs on Real Use Cases
Explore real-world DGL applications including GNN-FiLM, ARGO, GATNE for e-commerce recommendations, and EEG-GCNN for neurological disease diagnosis.DGL in Depth: SE(3)-Transformer on ROCm 7
Review this step-by-step implementation guide for benchmark results showing the great latency and throughput performance of the AMD Instinct MI300X GPU on SE(3)-Transformer workloads.
Prerequisites#
This tutorial was developed and tested using the following setup.
Hardware requirements#
GPU: AMD Instinct™ MI300X or MI250X GPUs.
Note: This tutorial has been tested and validated on AMD Instinct MI300X GPUs. For the official list of supported GPUs, see the DGL Compatibility Documentation. For performance metrics on AMD Instinct GPUs, see the SE(3)-Transformer performance report blog.
Software requirements#
AMD validates and publishes DGL images with ROCm backends on Docker Hub. The following table shows the validated software stack configurations:
Software |
Supported versions |
Notes |
|---|---|---|
ROCm |
6.4.0, 6.4.3, 7.0.0 |
AMD GPU compute platform |
Python |
3.10, 3.12 |
Programming language |
PyTorch |
2.3.0, 2.4.1, 2.6.0, 2.7.1, 2.8.0 |
Deep learning framework (ROCm build) |
DGL |
2.4.0 |
Deep Graph Library for GNNs |
Ubuntu |
22.04, 24.04 |
Operating system |
Docker |
20.10+ |
Container runtime (recommended) |
Validated Docker images#
The recommended approach is to use the prebuilt Docker images from AMD. Select the image that matches your desired configuration:
Docker image tag |
DGL release |
ROCm |
PyTorch |
Ubuntu |
Python |
|---|---|---|---|---|---|
|
25.10 |
7.0.0 |
2.8.0 |
24.04 |
3.12 |
|
25.10 |
7.0.0 |
2.6.0 |
24.04 |
3.12 |
|
25.10 |
7.0.0 |
2.7.1 |
22.04 |
3.10 |
|
25.10 |
6.4.3 |
2.6.0 |
24.04 |
3.12 |
|
25.07 |
6.4.0 |
2.6.0 |
24.04 |
3.12 |
|
25.07 |
6.4.0 |
2.4.1 |
24.04 |
3.12 |
|
25.07 |
6.4.0 |
2.4.1 |
22.04 |
3.10 |
|
25.07 |
6.4.0 |
2.3.0 |
22.04 |
3.10 |
Set up the SE(3)-Transformer environment#
In this tutorial, you will work on the prebuilt ROCm DGL image as an example. You can also use other validated DGL images from the table above.
Step 1: Launch the Docker image#
Launch the Docker container. Replace /path/to/workspace with the full path to the directory on your host machine where you want to clone the DeepLearningExamples code and run the notebook. Choose the image tag from the validated Docker images table above that matches your desired configuration.
docker run -it --rm --privileged \
--network=host \
--device=/dev/kfd \
--device=/dev/dri \
--group-add=video \
--ipc=host \
--cap-add=SYS_PTRACE \
--security-opt seccomp=unconfined \
-v /path/to/workspace:/workspace \
-w /workspace \
<IMAGE_TAG>
Note: This command mounts your host directory to /workspace in the container. Ensure the notebook file is in this directory or upload it after Jupyter starts. The remaining steps should be run inside the Docker container. Save the URL (and token, if shown) from the terminal output to access JupyterLab from your browser.
For example, to use ROCm 7.0.0 with PyTorch 2.7.1:
docker run -it --rm --privileged \
--network=host \
--device=/dev/kfd \
--device=/dev/dri \
--group-add=video \
--ipc=host \
--cap-add=SYS_PTRACE \
--security-opt seccomp=unconfined \
-v /path/to/workspace:/workspace \
-w /workspace \
rocm/dgl:dgl-2.4.0.amd0_rocm7.0.0_ubuntu22.04_py3.10_pytorch_2.7.1
Step 2: Install and launch Jupyter#
Inside the Docker container, install Jupyter and the visualization packages used by this notebook:
pip install jupyter
Start the Jupyter server:
jupyter-lab --ip=0.0.0.0 --port=8888 --no-browser --allow-root
Note: Ensure port 8888 is not already in use on your system before running the above command. If it is, you can specify a different port by replacing --port=8888 with another port number, for example, --port=8890.
Step 3: Open the notebook#
Once JupyterLab is running, open your browser and go to the URL shown in the terminal (typically http://localhost:8888). In the file browser, navigate to DGLPyTorch/DrugDiscovery/SE3Transformer/ and click se3transformer.ipynb (or this intro notebook) to begin. Alternatively, you can upload this notebook to your Jupyter lab via the upload button in Jupyter.
Two ways to use this notebook#
This notebook supports two modes:
Quick inference mode (~10 minutes)
Use a pretrained model trained for 100 epochs.
Skip training and go straight to inference and results.
Perfect for exploring model capabilities quickly.
Training mode (~30-60 minutes)
Train from scratch for five epochs.
See the full pipeline: data → model → train → evaluate.
Perfect for understanding the complete workflow.
To select a mode, set USE_PRETRAINED = True (for inference) or USE_PRETRAINED = False (for training) in the configuration cell below.
Note: Run the rest of this notebook in Jupyter by executing the cells.
Loading the dependencies and repository#
This section explains how to download and install the required code and dependencies.
Install dependencies#
!pip install plotly torchinfo rdkit py3Dmol
Clone the DeepLearningExamples repository#
Clone the ROCm DeepLearningExamples repository using sparse checkout to download only the SE(3)-Transformer code. Run the following inside the Docker container (for example, in a terminal or in a notebook code cell):
%%bash
git clone --filter=blob:none --sparse https://github.com/ROCm/DeepLearningExamples.git
cd DeepLearningExamples
git sparse-checkout set DGLPyTorch/DrugDiscovery/SE3Transformer
cd DGLPyTorch/DrugDiscovery/SE3Transformer
pip install -r requirements.txt
pip install -e .
Add installed packages to path#
Import the necessary packages and add them to the path.
import os
import sys
# Get current working directory and build the SE3 path
base_dir = os.getcwd()
se3_path = os.path.abspath(os.path.join(base_dir, "DeepLearningExamples/DGLPyTorch/DrugDiscovery/SE3Transformer"))
# Verify it exists and add to path
if os.path.exists(se3_path):
if se3_path not in sys.path:
sys.path.insert(0, se3_path)
print(f"✅ Added {se3_path} to Python path")
print(f" Current working directory: {base_dir}")
else:
print(f"❌ Error: Path not found: {se3_path}")
print(f" Please check if the repository was cloned correctly")
What happens next?#
First, choose between inference or training mode.
# ============================================
# 🎯 CONFIGURATION: Choose Your Path
# ============================================
USE_PRETRAINED = False # Toggle: True = Inference only | False = Train from scratch
if USE_PRETRAINED:
print("="*60)
print("📊 MODE: Quick Inference with Pretrained Model")
print("="*60)
print("✅ Will load: model_qm9_100_epochs.pth")
print("⏭️ Training cells will be skipped")
print("⚡ Estimated time: ~10 minutes\n")
CHECKPOINT_PATH = "model_qm9_100_epochs.pth"
EPOCHS = None
else:
print("="*60)
print("🏋️ MODE: Training from Scratch")
print("="*60)
print("✅ Will train for 5 epochs")
print("💾 Will save to: model_qm9_5_epochs.pth")
print("⏱️ Estimated time: ~30-60 minutes\n")
CHECKPOINT_PATH = "model_qm9_5_epochs.pth"
EPOCHS = 5
Depending on which setting you choose, the next steps vary somewhat:
If
USE_PRETRAINED = True(inference mode):Import the libraries
Load the dataset and inspect the molecules
Initialize the model architecture
Skip training, logging setup, and visualization
Load a pretrained checkpoint (
model_qm9_100_epochs.pth)Run inference and see the results
If
USE_PRETRAINED = False(training mode):Import the libraries
Load the dataset and inspect the molecules
Initialize the model architecture
Train for five epochs
Visualize training progress
Load your trained checkpoint (
model_qm9_5_epochs.pth)Run inference and see the results
Imports for training and evaluation#
These imported modules set up the full SE(3)-Transformer training and evaluation pipeline on the QM9 molecular dataset. They cover data loading, distributed training, optimization, logging, and inference.
# Core imports needed for both training and inference
import torch.nn as nn
import dgl
from se3_transformer.data_loading import QM9DataModule
from se3_transformer.model import SE3TransformerPooled
from se3_transformer.model.fiber import Fiber
from se3_transformer.runtime.arguments import PARSER
from se3_transformer.runtime.utils import (
seed_everything,
using_tensor_cores,
)
import logging
from se3_transformer.runtime.callbacks import (
QM9MetricCallback,
QM9LRSchedulerCallback,
)
from se3_transformer.runtime.loggers import (
LoggerCollection,
DLLogger,
)
from se3_transformer.runtime.training import train
print("✅ Loaded training modules")
Using the CLI arguments to set up training#
The SE(3)-Transformer example from the DeepLearningExamples repository was originally designed to run as a command-line program, but you can easily adapt it to use in a Jupyter notebook. The training configuration, including model, optimizer, and runtime settings, is managed through an argparse parser, which you can leverage directly within the notebook for flexible experimentation.
if not USE_PRETRAINED:
# Uncomment the line below to see all available training and runtime arguments
# PARSER.print_help()
# Training configuration - adjust these parameters as needed
TRAINING_ARGS = [
"--epochs", str(EPOCHS), # Number of training epochs (5 for quick training)
"--eval_interval", "1", # Evaluate every N epochs
"--batch_size", "240", # Batch size per GPU
"--num_workers", "16", # Data loader workers
"--precompute_bases", # Precompute geometric bases for speed
"--use_layer_norm", # Use layer normalization
"--norm", # Normalize features
"--save_ckpt_path", CHECKPOINT_PATH, # Where to save the trained model
"--task", "homo", # Prediction task (homo, lumo, gap, etc.)
"--amp", # Enable automatic mixed precision
"--learning_rate", "0.002", # Initial learning rate
"--weight_decay", "0.1", # L2 regularization
"--seed", "42", # Random seed for reproducibility
]
# Parse arguments
args = PARSER.parse_args(TRAINING_ARGS)
print("="*60)
print("🔧 Training Configuration")
print("="*60)
print(f"Epochs: {args.epochs}")
print(f"Batch Size: {args.batch_size}")
print(f"Learning Rate: {args.learning_rate}")
print(f"Weight Decay: {args.weight_decay}")
print(f"Checkpoint Path: {args.save_ckpt_path}")
print(f"Task: {args.task}")
print(f"AMP Enabled: {args.amp}")
print("="*60)
else:
# For pretrained mode, we still need args for model initialization
# but we won't use them for training
args = PARSER.parse_args([
"--epochs", "100",
"--batch_size", "240",
"--num_workers", "16",
"--use_layer_norm",
"--norm",
"--task", "homo",
])
print("⏭️ Skipping training configuration (using pretrained model)")
Download the pretrained model from Hugging Face#
If you are using pretrained mode (USE_PRETRAINED = True), the model_qm9_100_epochs.pth checkpoint must be available locally. This checkpoint is hosted on the Hugging Face Hub. To download it, follow these steps:
Install the Hugging Face Hub client using
pip install huggingface_hub.Log in to Hugging Face (recommended for gated or rate-limited access). Run
huggingface-cli loginin a terminal and enter your token from huggingface.co/settings/tokens.Download the model into the current directory with:
huggingface-cli download amd/se3_transformers model_qm9_100_epochs.pth --local-dir .
The cell below automatically runs these steps when USE_PRETRAINED is True.
if USE_PRETRAINED:
# Install Hugging Face Hub if not already installed
try:
from huggingface_hub import hf_hub_download
except ImportError:
import subprocess
import sys
subprocess.check_call([sys.executable, "-m", "pip", "install", "-q", "huggingface_hub"])
from huggingface_hub import hf_hub_download
# Advise logging in for gated repos or to avoid rate limits
print("💡 If the model is gated or you hit rate limits, log in first: huggingface-cli login")
print(" Get a token at: https://huggingface.co/settings/tokens\n")
# Download pretrained checkpoint to current directory
print("📥 Downloading model_qm9_100_epochs.pth from amd/se3_transformers ...")
path = hf_hub_download(
repo_id="amd/se3_transformers",
filename="model_qm9_100_epochs.pth",
local_dir=".",
local_dir_use_symlinks=False,
)
print(f"✅ Downloaded to: {path}")
else:
print("⏭️ Skipping download (training from scratch)")
Dataset and model setup#
Start by loading the QM9 molecular dataset using QM9DataModule, which handles data preprocessing, batching, and splitting for training and evaluation.
Next, initialize the SE(3)-Transformer model (SE3TransformerPooled) with input, edge, and output fibers that define how geometric and feature information flows through the network.
Finally, define the L1 loss (nn.L1Loss) — a simple yet effective choice for molecular property regression tasks.
datamodule = QM9DataModule(**vars(args))
model = SE3TransformerPooled(
fiber_in=Fiber({0: datamodule.NODE_FEATURE_DIM}),
fiber_out=Fiber({0: args.num_degrees * args.num_channels}),
fiber_edge=Fiber({0: datamodule.EDGE_FEATURE_DIM}),
output_dim=1,
tensor_cores=using_tensor_cores(args.amp), # use Tensor Cores more effectively
**vars(args),
)
loss_fn = nn.L1Loss()
Inspecting the molecules#
Before diving into training, it’s helpful to visually inspect the molecules from the QM9 dataset.
Review one of the molecules in the dataset:
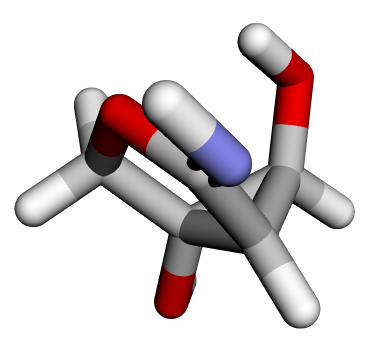
Basic graph information#
--- BASIC INFO ---
Nodes: 14
Edges: 28
The molecule is represented as a graph with 14 nodes corresponding to atoms and 28 edges representing atom–atom interactions. Edges are constructed based on interatomic proximity rather than explicit chemical bonds.
Node features (ndata)#
Each node (atom) is associated with geometric and chemical features.
Atomic positions#
Key: pos
Shape: (14, 3)
Dtype: torch.float32
tensor([[ 0.6781, -0.0583, 0.7324],
[-0.1432, 0.3297, -0.3889],
[-1.5010, -0.2886, -0.2937],
...])
Each row represents the 3D Cartesian coordinates [x,y,z] of an atom in the molecule.
Atomic attributes#
Key: attr
Shape: (14, 11)
Dtype: torch.float32
tensor([[0., 0., 0., 1., 0., 8., 0., 0., 0., 0., 1.],
[0., 1., 0., 0., 0., 6., 0., 0., 0., 0., 2.],
[0., 1., 0., 0., 0., 6., 0., 0., 0., 0., 0.],
...])
Each row encodes atom-specific properties, such as atomic type and related categorical or numerical descriptors, which allow the model to distinguish between different elements.
Edge features (edata)#
Edges capture pairwise relationships between atoms.
Edge Attributes
Key: edge_attr
Shape: (28, 4)
Dtype: torch.float32
tensor([[1., 0., 0., 0.],
[1., 0., 0., 0.],
[1., 0., 0., 0.],
...])
Each row represents a feature vector associated with an edge, typically encoding distance-based or radial information used to model interatomic interactions.
RAW values of HOMO, LUMO, and GAP#
The raw values of HOMO, LUMO and GAP in eV is as follows.
HOMO (Highest Occupied Molecular Orbital) energy#
Shape: torch.Size([1])
Value: tensor([-5.7987]) eV
LUMO (Lowest Unoccupied Molecular Orbital) energy#
Shape: torch.Size([1])
Value: tensor([0.9905]) eV
GAP (gap between HOMO and LUMO)#
Shape: torch.Size([1])
Value: tensor([6.7892]) eV
View the model summary#
Quickly inspect the SE(3)-Transformer architecture using torchinfo.summary, which prints a detailed overview of each layer, its input/output shapes, and the number of parameters. This helps you verify that the model has been built correctly before training.
from torchinfo import summary
summary(model)
Part A: Training pipeline (optional)#
Use this code to configure the training pipeline.
Note: If USE_PRETRAINED is set to True, the training cells below will be skipped, and you’ll proceed directly to inference.
Logging and callbacks#
# Initialize logging, set seed, configure loggers and training callbacks
logging.getLogger().setLevel(logging.INFO)
if args.seed is not None:
logging.info(f"Using seed {args.seed}")
seed_everything(args.seed)
logging.info(f"Saving info to {args.log_dir}/{args.dllogger_name}")
loggers = [DLLogger(save_dir=args.log_dir, filename=args.dllogger_name)]
logger = LoggerCollection(loggers)
callbacks = [
QM9MetricCallback(logger, targets_std=datamodule.targets_std, prefix="validation"),
QM9LRSchedulerCallback(logger, epochs=args.epochs),
]
logger.log_hyperparams(vars(args))
Train#
With everything configured, you’re ready to kick off training. The train() function orchestrates the entire training loop — running forward and backward passes, computing losses, updating parameters, and periodically evaluating the validation set. It uses the dataloaders, callbacks, and logger you set up earlier to track progress, log metrics, and manage learning rate schedules throughout the training process.
if not USE_PRETRAINED:
train(
model,
loss_fn,
datamodule.train_dataloader(),
datamodule.val_dataloader(),
callbacks,
logger,
args,
)
else:
print("⏭️ Skipping training (USE_PRETRAINED=True)")
print("📊 Will load pretrained checkpoint for inference")
Visualizing the training progress#
After training, you can visualize and analyze the logged results. Import Plotly for interactive plotting and dllogger to access the saved training logs. Flushing the logger ensures all metrics have been written to disk before loading them.
if not USE_PRETRAINED:
import plotly.graph_objects as go
import plotly.io as pio
import pandas as pd
from plotly.subplots import make_subplots
import json
import dllogger
import os
# If we're loading a checkpoint, we need to use the saved log file
# otherwise, we'll use the current log file
if args.load_ckpt_path is not None:
LOG_FILE = os.path.join("results", "dllogger_results_100.json")
if not os.path.exists(LOG_FILE):
raise FileNotFoundError(f"Log file {LOG_FILE} does not exist, please copy the log file to the results directory or turn off checkpoint loading")
else:
LOG_FILE = os.path.join("results", args.dllogger_name)
dllogger.flush()
print(f"Using log file: {LOG_FILE}")
pio.renderers.default = "notebook"
else:
print("⏭️ Skipping training visualization (no training was performed)")
This step parses and organizes the logged training data from dllogger_results.json. It reads the file line by line, cleans up any malformed entries, and filters out records without valid steps. Each log entry is then grouped by its training step, extracting key metrics such as training loss, learning rate, and validation mean absolute error (MAE). The results are compiled into a tidy Pandas DataFrame, making it easier to visualize and analyze how model performance and learning dynamics evolved throughout training.
if not USE_PRETRAINED:
# Read and parse the data
with open(LOG_FILE, "r") as f:
logs = [json.loads(line.replace("DLLL", "")) for line in f.readlines()]
# Filter out entries where step is an empty list
logs = [log for log in logs if log.get("step") != []]
# Create a dictionary to aggregate metrics by step
metrics_by_step = {}
for log in logs:
if log.get("type") == "LOG":
step = log.get("step")
# Skip if step is not an integer or if it's the PARAMETER step
if not isinstance(step, int):
continue
# Initialize the step if not exists
if step not in metrics_by_step:
metrics_by_step[step] = {
"step": step,
"train loss": None,
"learning rate": None,
"validation MAE": None,
}
# Update metrics for this step
data = log.get("data", {})
if "train loss" in data:
metrics_by_step[step]["train loss"] = data["train loss"]
if "learning rate" in data:
metrics_by_step[step]["learning rate"] = data["learning rate"]
if "validation MAE" in data:
metrics_by_step[step]["validation MAE"] = data["validation MAE"]
# Convert to DataFrame
df = pd.DataFrame(list(metrics_by_step.values()))
df = df.sort_values("step").reset_index(drop=True)
print(df)
else:
print("⏭️ Skipping training visualization (no training was performed)")
To get a clear picture of how training evolved, plot the key metrics over epochs using Plotly. The figure below displays training loss, validation MAE, and learning rate in separate subplots, making it easy to observe the model’s convergence and learning dynamics. Ideally, you should see the training loss and validation MAE steadily decreasing as the learning rate adjusts, giving quick visual confirmation that training progressed smoothly.
if not USE_PRETRAINED:
# Create subplots
fig = make_subplots(
rows=3,
cols=1,
subplot_titles=("Train Loss", "Validation MAE", "Learning Rate"),
vertical_spacing=0.08,
)
# Train Loss
fig.add_trace(
go.Scatter(
x=df["step"],
y=df["train loss"],
mode="lines+markers",
name="Train Loss",
line=dict(color="blue"),
),
row=1,
col=1,
)
# Validation MAE
fig.add_trace(
go.Scatter(
x=df["step"],
y=df["validation MAE"],
mode="lines+markers",
name="Validation MAE",
line=dict(color="red"),
),
row=2,
col=1,
)
# Learning Rate
fig.add_trace(
go.Scatter(
x=df["step"],
y=df["learning rate"],
mode="lines+markers",
name="Learning Rate",
line=dict(color="green"),
),
row=3,
col=1,
)
fig.update_xaxes(title_text="Epoch", row=3, col=1)
fig.update_layout(height=1000, showlegend=False, title_text="SE(3) Training")
fig.show()
else:
print("⏭️ Skipping training visualization (no training was performed)")
Here’s an example of what the output graph might look like:
Part B: Inference and evaluation#
This section loads a trained model checkpoint and evaluates it on the test set.
If
USE_PRETRAINED = True: Loads the 100-epoch pretrained model.If
USE_PRETRAINED = False: Loads the model you just trained for five epochs.
Set up inference configuration#
For inference, the model runs a forward-only pass. This means no gradients are computed, and the focus is on making predictions rather than updating weights.
import torch
# Get the major and minor compute capability of the current CUDA device
major_cc, minor_cc = torch.cuda.get_device_capability()
print(f"CUDA Compute Capability: {major_cc}.{minor_cc}")
Set up inference arguments#
If your SE(3)-Transformer code uses argparse to manage configurations, you can simulate command-line arguments in the notebook:
args_inference = PARSER.parse_args([
"--amp", # Enable automatic mixed precision (faster inference)
"true",
"--batch_size", # Number of molecules to process at once
"240",
"--use_layer_norm", # Enable layer normalization
"--norm", # Use normalization in the model
"--load_ckpt_path", # Path to the trained model checkpoint
CHECKPOINT_PATH,
"--task", # Prediction task (e.g., HOMO/LUMO energies)
"homo",
])
Get local GPU info#
Before running inference, check the GPU and prepare the device-specific settings:
from se3_transformer.runtime.utils import init_distributed, get_local_rank
# Initialize distributed utilities (still works for single-GPU)
is_distributed = init_distributed() # False for single-GPU
local_rank = get_local_rank() # GPU index, usually 0
print(f"Running on GPU: {local_rank}, Distributed: {is_distributed}")
Initialize the model#
Create the SE3Transformer for inference:
from se3_transformer.model import SE3TransformerPooled, Fiber
model = SE3TransformerPooled(
fiber_in=Fiber({0: datamodule.NODE_FEATURE_DIM}), # Node feature dimensions
fiber_out=Fiber({0: args.num_degrees * args.num_channels}), # Output fiber dimensions
fiber_edge=Fiber({0: datamodule.EDGE_FEATURE_DIM}), # Edge features
output_dim=1, # Single target prediction
tensor_cores=(args.amp and major_cc >= 7) or major_cc >= 8, # Use tensor cores if available
**vars(args) # Other parser arguments
)
Set up evaluation callbacks#
This computes relevant QM9 metrics during inference and uses dataset normalization to scale predictions properly.
callbacks = [
QM9MetricCallback(logger, targets_std=datamodule.targets_std, prefix='test'),
QM9LRSchedulerCallback(logger, epochs=args.epochs),
]
Load the pretrained checkpoint#
The following code loads the checkpoint you previously downloaded.
print(f"📂 Loading checkpoint: {CHECKPOINT_PATH}")
import torch
checkpoint = torch.load(
str(args_inference.load_ckpt_path),
map_location=f'cuda:{local_rank}', # Map weights to the active GPU
weights_only=True # Only load model parameters
)
model.load_state_dict(checkpoint['state_dict'])
torch.set_float32_matmul_precision('high')
test_dataloader = datamodule.test_dataloader()
device = torch.cuda.current_device()
model.to(device)
After this step, the model is ready for inference.
Running evaluation on the test set#
After the model is loaded and ready, you can run evaluation on the test dataset.
from se3_transformer.runtime.training import evaluate
# Run the evaluation function
evaluate(model, test_dataloader, callbacks, args_inference)
# Trigger the 'on_validation_end' hook for all callbacks
for callback in callbacks:
callback.on_validation_end()
Post-inference analysis#
Referring to the previous example, you can now inspect the molecule, examine its regression targets, and compare it with the prediction.

To obtain these values, run separate training and inference sessions for each task, specifying the --task argument as homo, lumo, or gap.
HOMO (Highest Occupied Molecular Orbital) energy
TARGET: tensor([1.2334])
PREDICTION: tensor([1.2236], dtype=torch.float16)
LUMO (Lowest Unoccupied Molecular Orbital) energy
TARGET : tensor([0.5246])
PREDICTION : tensor([0.5127], dtype=torch.float16)
Gap (Energy difference between HOMO and LUMO)
TARGET: tensor([-0.0547])
PRED: tensor([-0.1097], dtype=torch.float16)
Conclusion#
This notebook walked you through the end-to-end workflow for training and evaluating an SE(3)-Transformer model on the QM9 molecular dataset. You explored how to set up training configurations originally designed for CLI use, adapted them for an interactive Jupyter workflow, and visualized molecules directly from graph data to validate preprocessing. You then built and trained the SE(3)-Transformer, logged its performance, and used interactive plots to analyze key metrics like loss, MAE, and learning rate over time.
With the workflow now validated, this setup provides a strong foundation for scaling up experiments, benchmarking performance, and adapting the SE(3)-Transformer to more complex or domain-specific datasets.